BLD Insights
Discussion on Modified Nucleosides
15 March 2022
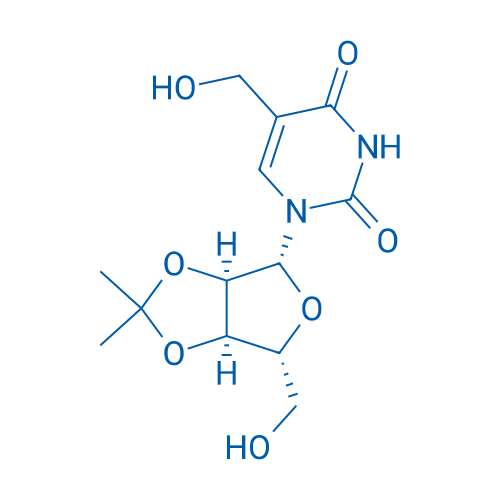
5-(Hydroxymethyl)-1-((3aR,4R,6R,6aR)-6-(hydroxymethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)pyrimidine-2,4(1H,3H)-dione
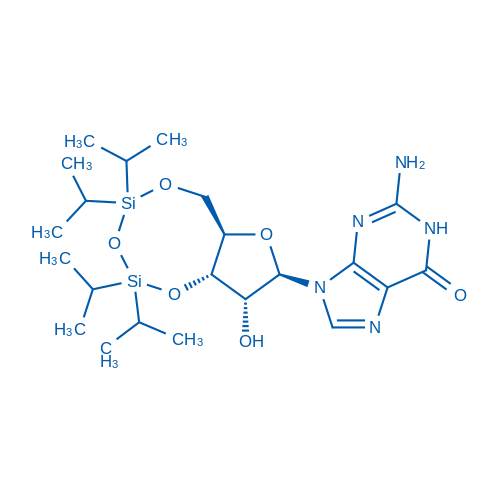
3',5'-O-(1,1,3,3-TEtraisopropyl-1,3-disiloxanediyl)guanosine
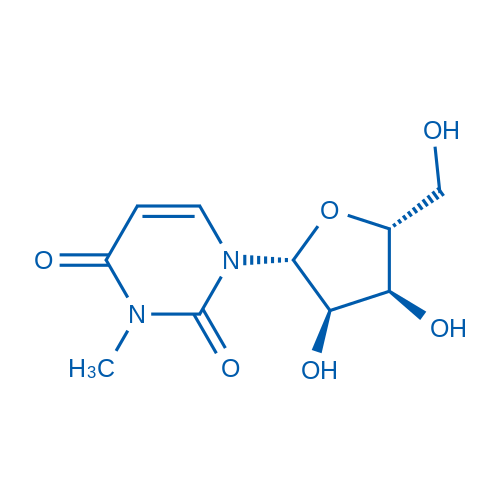
1-((2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-3-methylpyrimidine-2,4(1H,3H)-dione
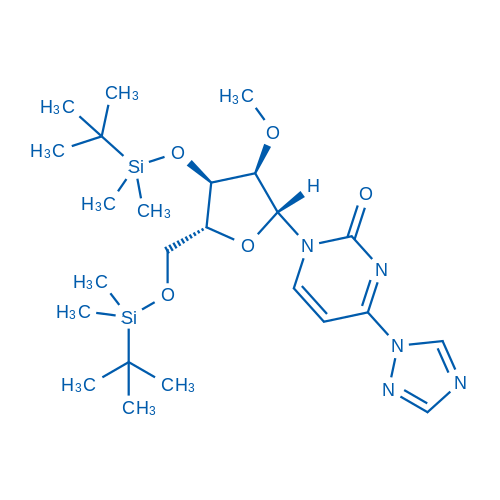
1-((2R,3R,4R,5R)-4-((tert-Butyldimethylsilyl)oxy)-5-(((tert-butyldimethylsilyl)oxy)methyl)-3-methoxytetrahydrofuran-2-yl)-4-(1H-1,2,4-triazol-1-yl)pyrimidin-2(1H)-one
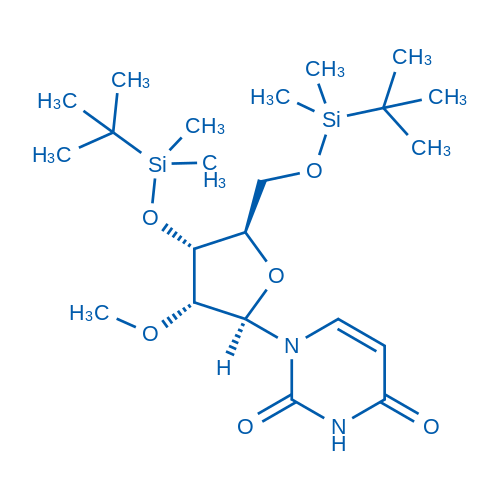
1-((2R,3R,4R,5R)-4-((tert-Butyldimethylsilyl)oxy)-5-(((tert-butyldimethylsilyl)oxy)methyl)-3-methoxytetrahydrofuran-2-yl)pyrimidine-2,4(1H,3H)-dione
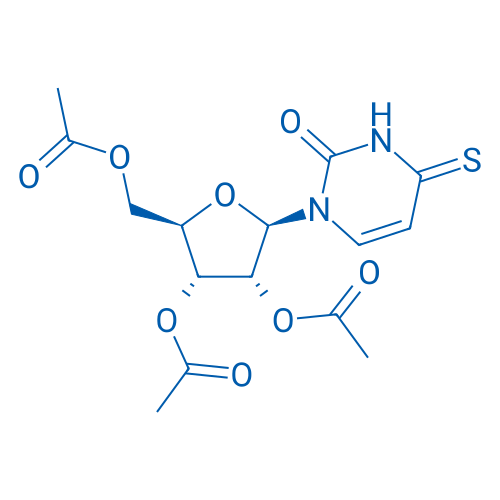
(2R,3R,4R,5R)-2-(Acetoxymethyl)-5-(2-oxo-4-thioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3,4-diyl diacetate
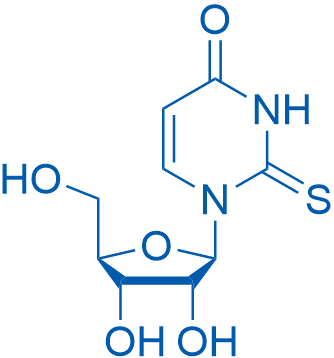
1-((2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2-thioxo-2,3-dihydropyrimidin-4(1H)-one
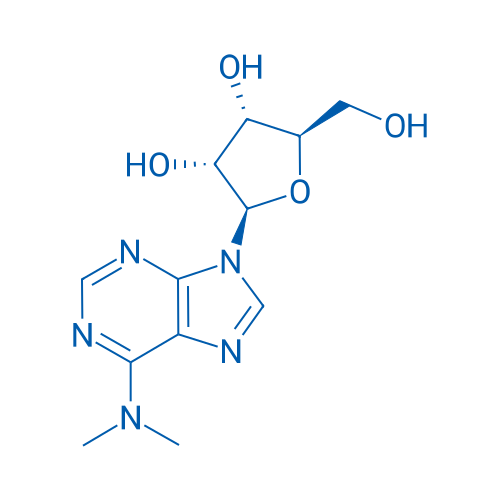
(2R,3R,4S,5R)-2-(6-(Dimethylamino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol
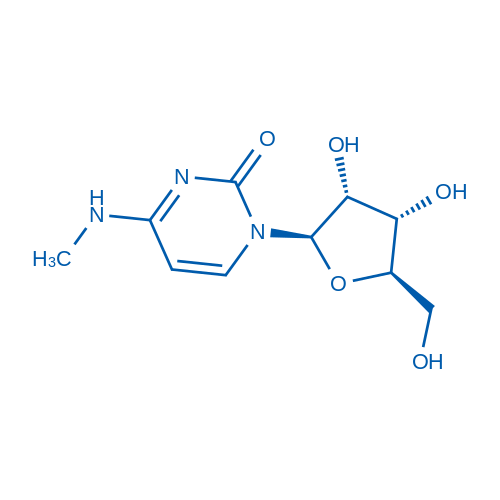
N4-Methylcytidine
Nucleosides are important raw materials for DNA and RNA in organisms and play a key role in various cellular processes ranging from cell signaling to cell proliferation. There are 8 kinds of natural nucleosides (Fig.1) and other modified nucleosides in cells (Fig.2). 1 Related research on modified nucleosides is advancing to explore the function and mechanism of modified nucleosides in organisms. And the study achievements are widely used in medicinal chemistry chemical biology. 2-5
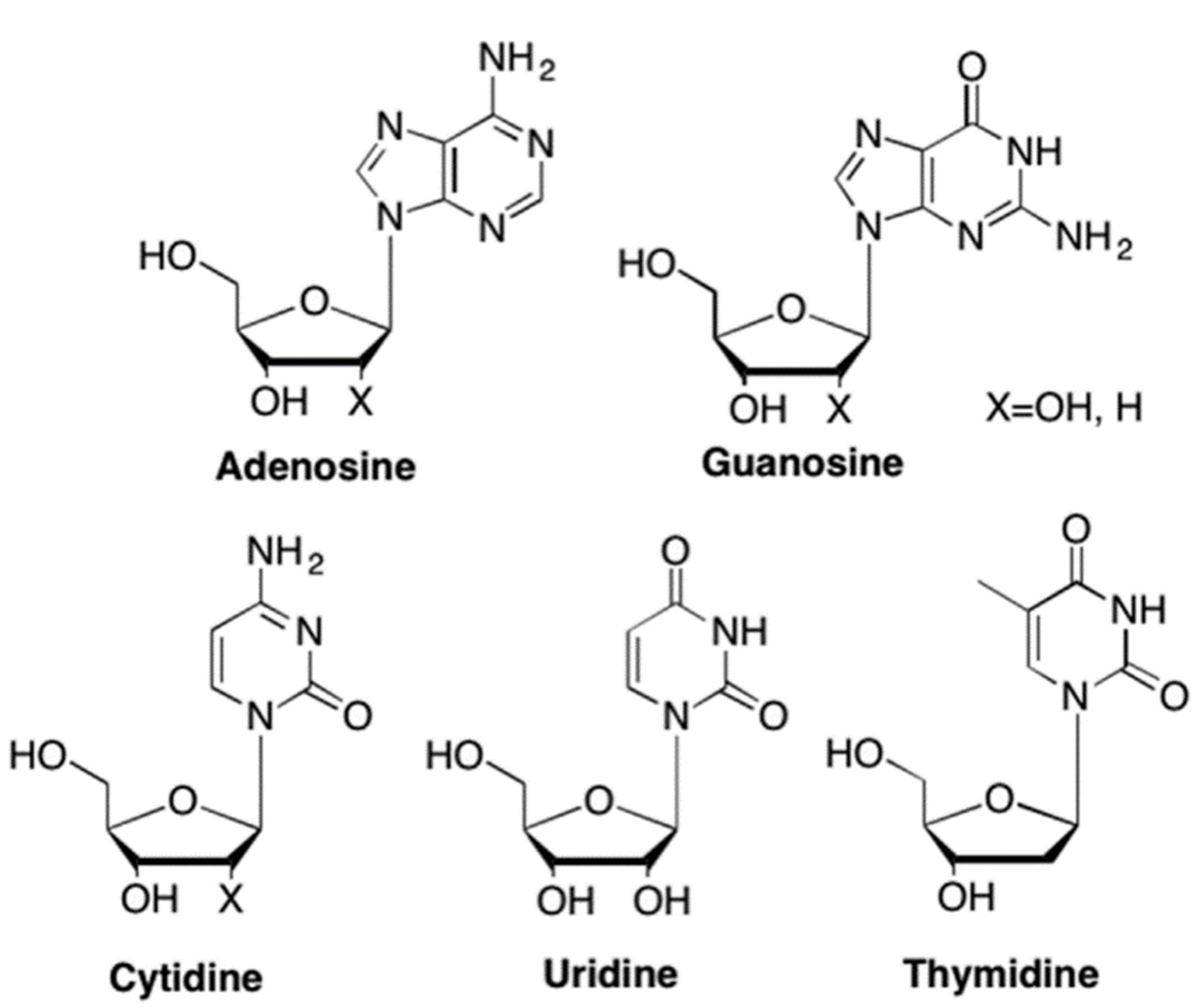
Fig.1: The most common nucleosides in RNA and DNA 6
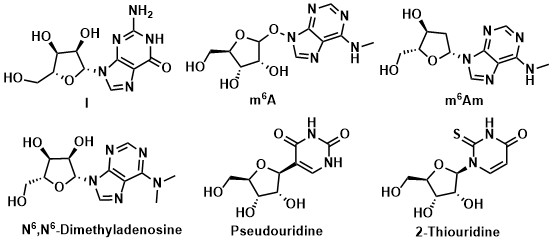
Fig.2: Naturally modified nucleosides in organisms
Mechanism
Modified nucleosides are phosphorylated in cells and will be recognized by host cells or viral enzymes to play a bioactive role: (1) They will compete with normal nucleosides and inhibit the growth of cancer cells, the replication of virus and other disease processes. (2) Triphosphate modified nucleotides can replace natural nucleotides and adjust the fate or function of DNA or RNA by changing base pairing, Π-Π stacking, or metal chelation. 7, 8 Due to the importance of DNA and RNA replication in rapidly proliferating tumor cells and virus replication, modified nucleosides can be used as drug molecules to treat diseases such as cancer and viral infections (Fig.3). 7, 9
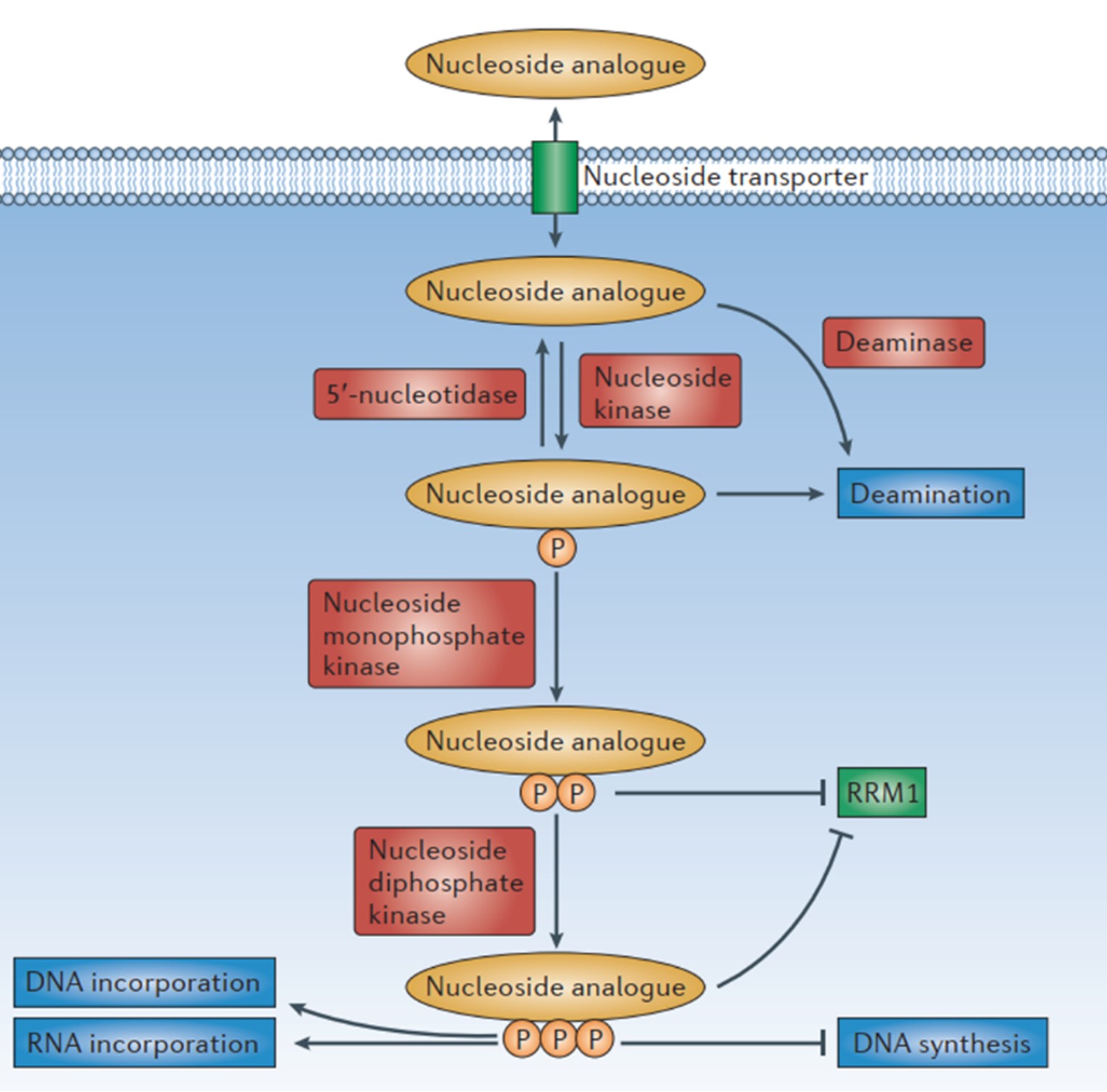
Fig.3: The mechanism of modified nucleosides in the cell. Cellular uptake of nucleoside analogues is an active process involving concentrative nucleoside transporters (CNTs) and equilibrative nucleoside transporters (ENTs). Modified nucleoside undergoes an initial rate-limiting phosphorylation step by a nucleoside kinase, which leads to the production of a monophosphate metabolite. A second phosphorylation step is then performed by nucleoside monophosphate kinase, and the third phosphorylation step is performed by nucleoside diphosphate kinase. Triphosphates can be incorporated in nucleic acids, in competition with their normal counterparts.
Optimization Strategy
Modified nucleosides are mainly used in the research and development of the following types of drugs (1) Anticancer drugs: cytarabine, azacitidine, gemcitabine, etc.(2) rheumatism drugs: azathioprine, allopurinol, etc.(3) antiviral drugs: acyclovir, monupivir, etc.(4) Antibiotics: trimethoprim, capillin, polyoxin J, etc.(5) Raw materials for nucleic acid drugs (such as DNA/RNA vaccines).4, 6, 10-17
In the research and development of related drugs, modification of nucleoside bases, glycosidic bonds and sugar rings is a common strategy to improve the pharmacokinetic and pharmacodynamic properties, reduce the immunogenicity of nucleic acid drugs and improve the stability: (1) Modification of sugar ring: PSI-6206 introduce substituents on the C2-5 position of furanose. It can improve metabolic stability and affect the conformation of furanose. The hydroxyl group of Molnupiravir is blocked with an acyl group to make a prodrug, which can effectively improve its cell membrane permeability. (2) Unnatural base modification: Modification of nucleosides based can also obtain better bioactivity and drug-like properties (such as Ribavirin). 18 (3) Change the connection mode of glycosidic bonds: such as changing the C-N bond to C-C/C-O/C-S and other connection methods can improve its metabolic properties (Fig.4).
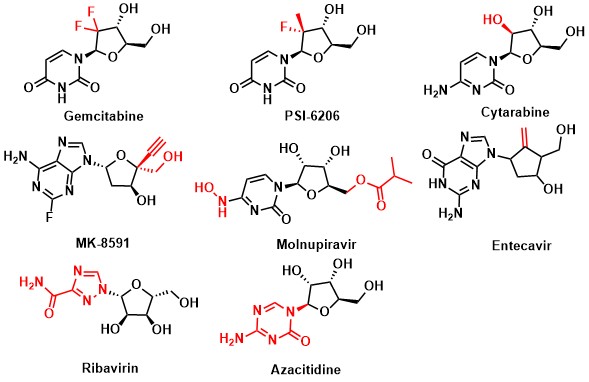
Fig.4: Approved drugs or preclinical drug candidates depended on modified nucleosides
Other Applications
Modified nucleosides are widely used in other aspects, such as the diagnosis of disease. With reporter groups (such as isotopic labels or fluorophores) or functional groups (such as azide, alkynyl, etc.), we could achieve the real-time localization or quantitative detection of nucleic acids. And it can be used to explore the structure, function, and fate of nucleic acids under physiological conditions (FIG.5, 6). (such as nucleic acid-protein interactions, nucleic acid structure, kinetic characteristics, etc.) 3, 19-22
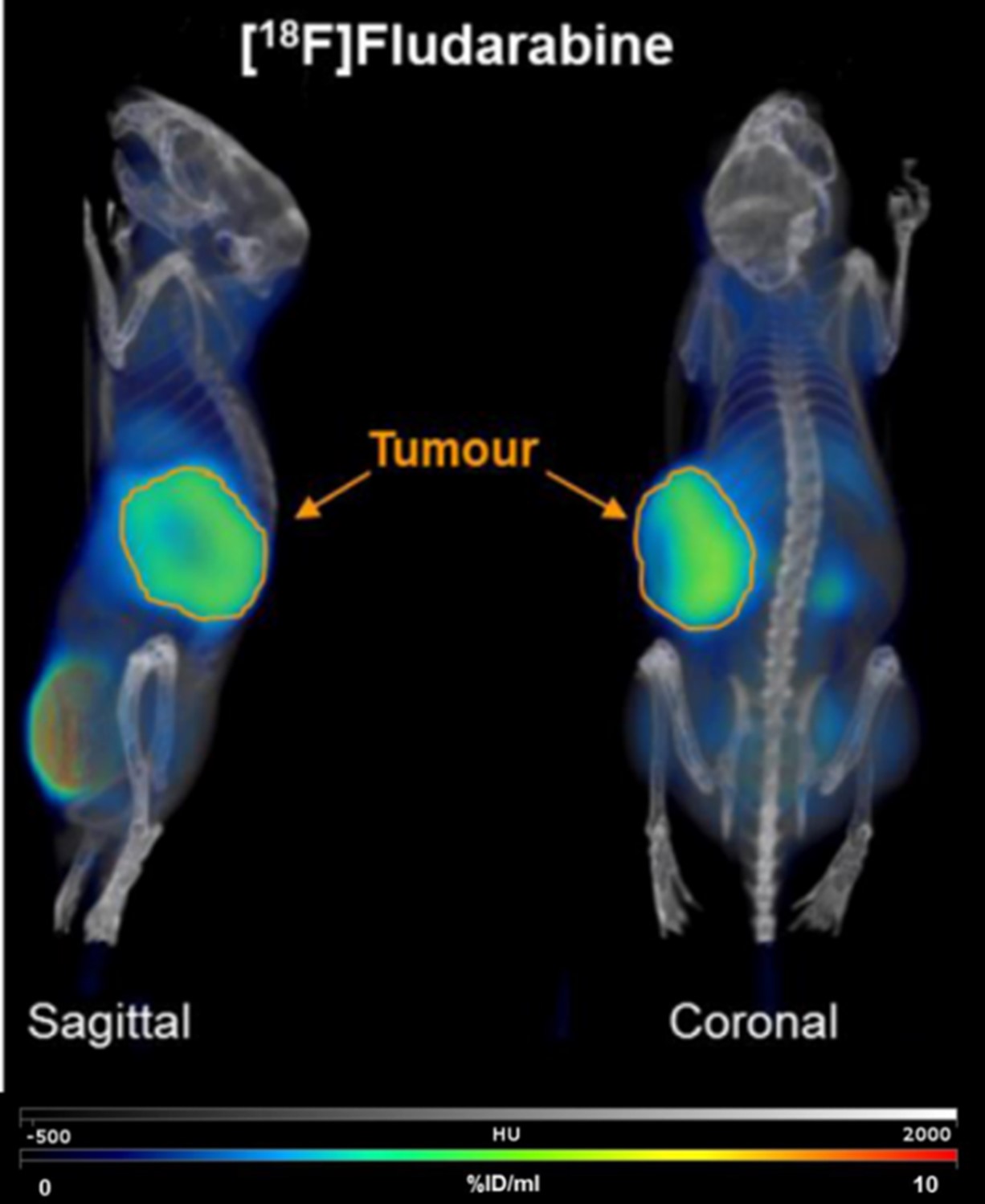
Fig.5: Imagining of [18F]-Fludarabine in a xenograft model
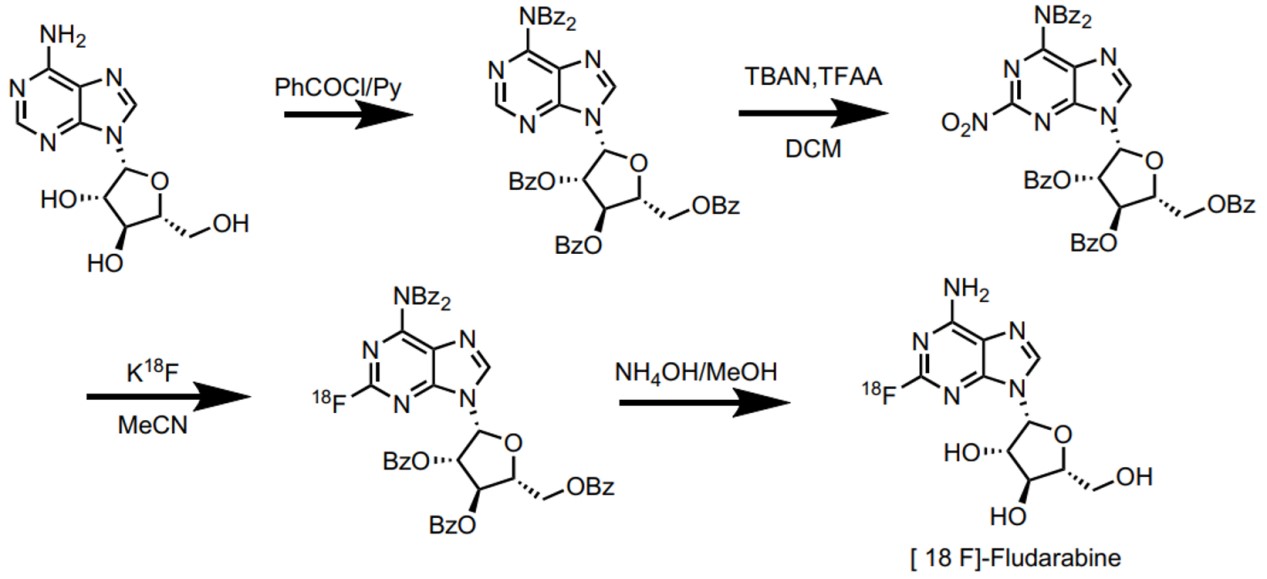
Fig.6: Synthesis route of [18F]-Fludarabine
References
[1]Zheng YY, Wu Y, Begley TJ, Sheng J. Sulfur modification in natural RNA and therapeutic oligonucleotides. RSC Chem Biol. 2021 Apr 27;2(4):990-1003.
[2]Pruijssers, A. J.; Denison, M. R., Nucleoside analogues for the treatment of coronavirus infections. Current Opinion in Virology 2019, 35, 57-62.
[3]Palumbo, C. M.; Beal, P. A., Nucleoside analogs in the study of the epitranscriptome. Methods 2019, 156, 46-52.
[4]Konopleva, M.; Letai, A., BCL-2 inhibition in AML: an unexpected bonus? Blood 2018, 132, 1007-1012.
[5]Tian, L.; Qiang, T.; Liang, C.; Ren, X.; Jia, M.; Zhang, J.; Li, J.; Wan, M.; YuWen, X.; Li, H.; Cao, W.; Liu, H., RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. European Journal of Medicinal Chemistry 2021, 213, 113201.
[6]Seley-Radtke, K. L.; Yates, M. K., The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antiviral Research 2018, 154, 66-86.
[7]Jordheim, L. P.; Durantel, D.; Zoulim, F.; Dumontet, C., Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nature Reviews Drug Discovery 2013, 12, 447-464.
[8]Meanwell, M.; Silverman Steven, M.; Lehmann, J.; Adluri, B.; Wang, Y.; Cohen, R.; Campeau, L.-C.; Britton, R., A short de novo synthesis of nucleoside analogs. Science 2020, 369, 725-730.
[9]Brocato, R. L.; Hooper, J. W., Progress on the Prevention and Treatment of Hantavirus Disease. Viruses 2019, 11.
[10]Imran, M.; Kumar Arora, M.; Asdaq, S. M.; Khan, S. A.; Alaqel, S. I.; Alshammari, M. K.; Alshehri, M. M.; Alshrari, A. S.; Mateq Ali, A.; Al-shammeri, A. M.; Alhazmi, B. D.; Harshan, A. A.; Alam, M. T.; Abida, Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19. Molecules 2021, 26.
[11]He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G., Functions of N6-methyladenosine and its role in cancer. Molecular Cancer 2019, 18, 176.
[12]Celik, I.; Erol, M.; Duzgun, Z., In silico evaluation of potential inhibitory activity of remdesivir, favipiravir, ribavirin and galidesivir active forms on SARS-CoV-2 RNA polymerase. Molecular Diversity 2021.
[13]Ogawa, E.; Wei, M. T.; Nguyen, M. H., Hepatitis B Virus Reactivation Potentiated by Biologics. Infectious Disease Clinics of North America 2020, 34, 341-358.
[14]Deeks, E. D., Bictegravir/Emtricitabine/Tenofovir Alafenamide: A Review in HIV-1 Infection. Drugs 2018, 78, 1817-1828.
[15]Niu, G.; Li, Z.; Huang, P.; Tan, H., Engineering nucleoside antibiotics toward the development of novel antimicrobial agents. The Journal of Antibiotics 2019, 72, 906-912.
[16]Yasui, H.; Iizuka, D.; Hiraoka, W.; Kuwabara, M.; Matsuda, A.; Inanami, O., Nucleoside analogs as a radiosensitizer modulating DNA repair, cell cycle checkpoints, and apoptosis. Nucleosides, Nucleotides & Nucleic Acids 2020, 39, 439-452.
[17]Bugg, T. D. H.; Kerr, R. V., Mechanism of action of nucleoside antibacterial natural product antibiotics. The Journal of Antibiotics 2019, 72, 865-876.
[18]Aher, U. P.; Srivastava, D.; Singh, G. P.; S, J. B., Synthetic strategies toward 1,3-oxathiolane nucleoside analogues. Beilstein Journal of Organic Chemistry 2021, 17, 2680-2715.
[19]Dziuba, D.; Didier, P.; Ciaco, S.; Barth, A.; Seidel, C. A. M.; Mély, Y., Fundamental photophysics of isomorphic and expanded fluorescent nucleoside analogues. Chemical Society Reviews 2021, 50, 7062-7107.
[20]Balasubramaniyam, T.; Oh, K.-I.; Jin, H.-S.; Ahn, H.-B.; Kim, B.-S.; Lee, J.-H., Non-Canonical Helical Structure of Nucleic Acids Containing Base-Modified Nucleotides. International Journal of Molecular Sciences 2021, 22.
[21]Kabza, A. M.; Kundu, N.; Zhong, W.; Sczepanski, J. T., Integration of chemically modified nucleotides with DNA strand displacement reactions for applications in living systems. WIREs Nanomedicine and Nanobiotechnology 2022, 14, e1743.
[22]Hovhannisyan, N.; Guillouet, S.; Fillesoye, F.; Dhilly, M.; Patin, D.; Galateau, F.; Leporrier, M.; Barré, L., Evaluation of the specificity of [18F]fludarabine PET/CT in a xenograft model of follicular lymphoma: comparison with [18F]FDG and impact of rituximab therapy. EJNMMI Research 2015, 5, 23.