BLD Insights
E3 Ligands Usage in PROTAC Design
19 April 2022
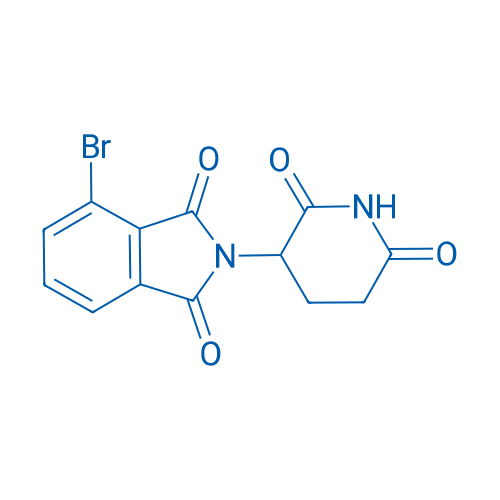
4-Bromo-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione
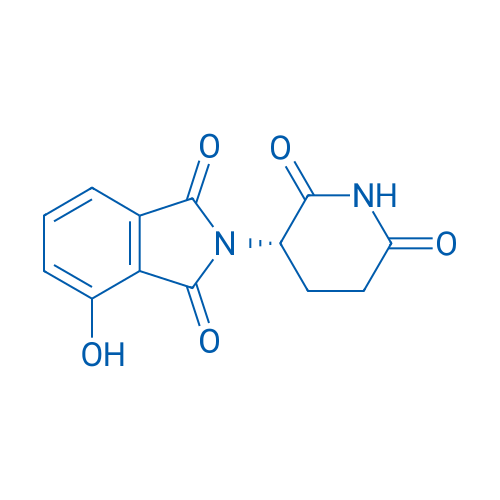
(S)-2-(2,6-Dioxopiperidin-3-yl)-4-hydroxyisoindoline-1,3-dione
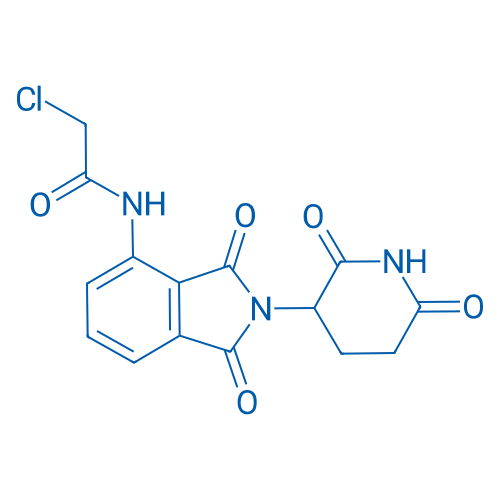
2-Chloro-N-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)acetamide
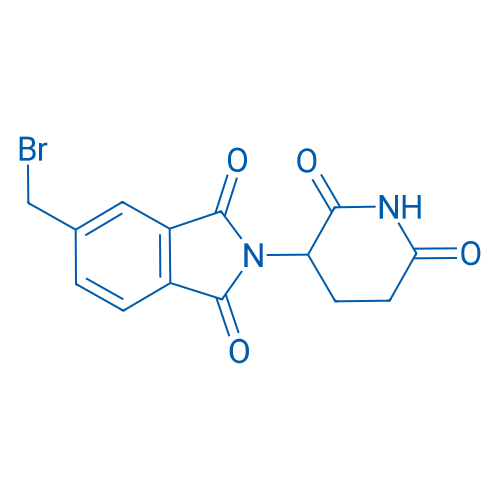
5-(Bromomethyl)-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione
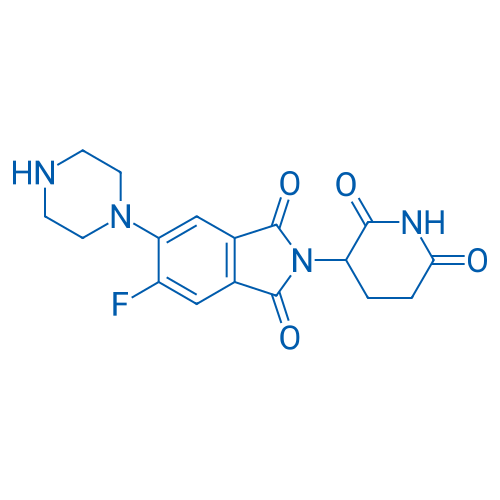
2-(2,6-Dioxopiperidin-3-yl)-5-fluoro-6-(piperazin-1-yl)isoindoline-1,3-dione
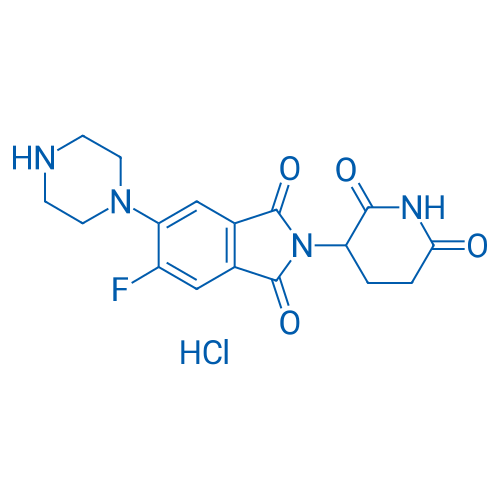
2-(2,6-Dioxopiperidin-3-yl)-5-fluoro-6-(piperazin-1-yl)isoindoline-1,3-dione hydrochloride
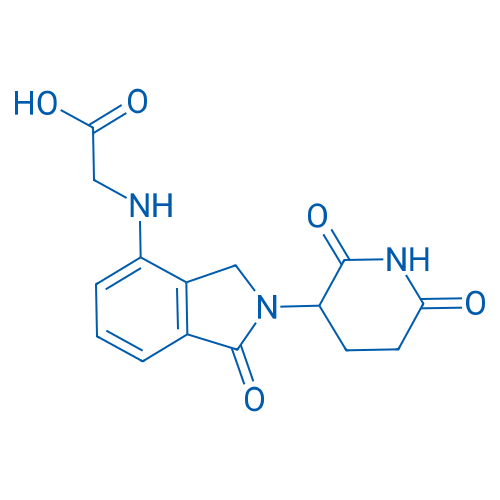
2-((2-(2,6-Dioxopiperidin-3-yl)-1-oxoisoindolin-4-yl)amino)acetic acid
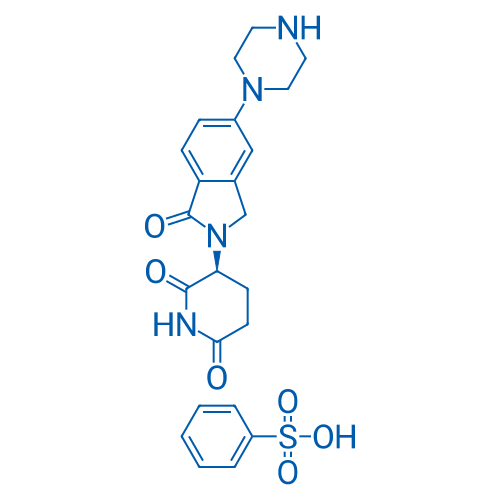
(S)-3-(1-Oxo-5-(piperazin-1-yl)isoindolin-2-yl)piperidine-2,6-dione benzenesulfonate
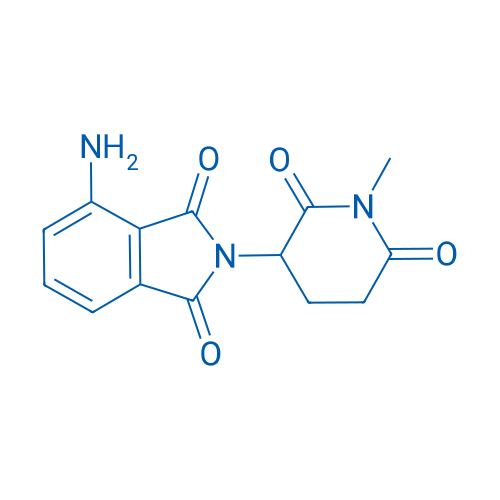
4-Amino-2-(1-methyl-2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione
Target protein degradation is a powerful strategy to modulate protein function and study the corresponding cell signaling pathways, including the use of proteolysis targeting chimeras (PROTACs) and molecular glue. 1 PROTACs is a bifunctional molecules to induce the proximity of target proteins to E3 ubiquitin ligases and utilize the ubiquitin-proteasome system to degrade proteins. E3 ligases have specific expression in a certain tissue type, cell type or cell compartment region. So their selective and specific ligands are important cornerstones for designing and expanding the chemical space of PROTAC and the range of target proteins provides assistance for the research and treatment of related diseases. 2 However, compared with the more than 600 E3 ligases encoded by the human genome, the number of E3 ligase ligands is limited (about 10~20 E3 ligases have corresponding ligands).Hence,further research on new E3 ligands is needed. 3-5
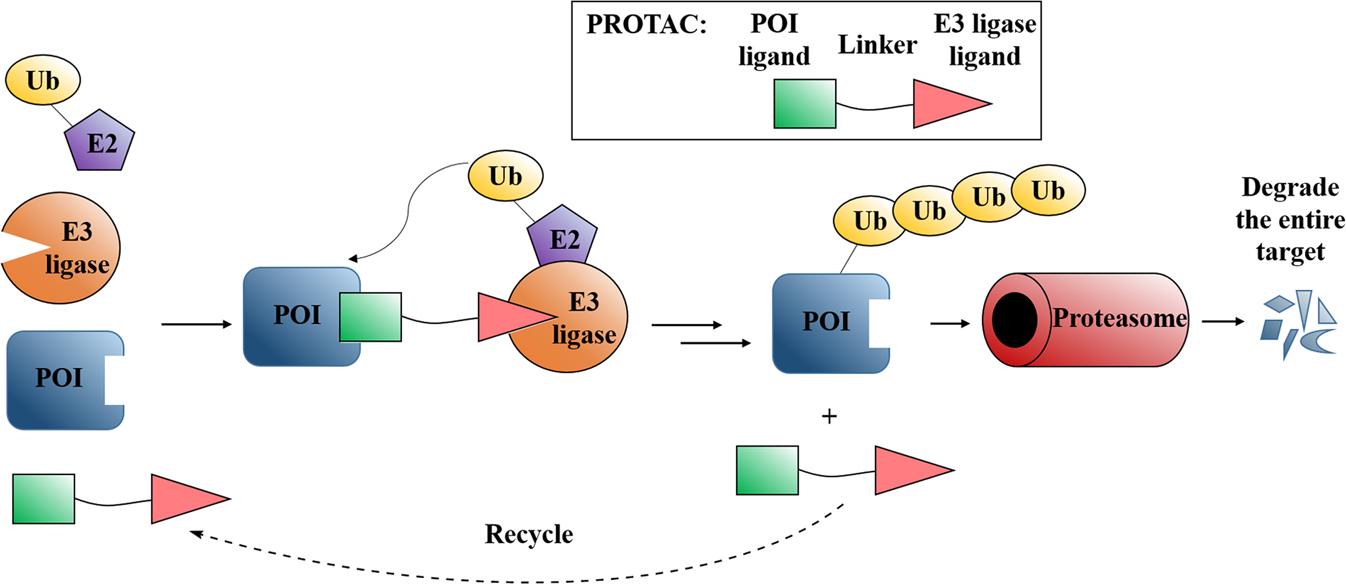
FIG 1: Mechanism and design strategy of PROTACs. The ubiquitin-proteasome system (UPS) is an important pathway for selective protein degradation. Under the action of a series of special enzymes such as ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin ligase (E3), ubiquitin molecules specifically modify target proteins to form target protein &polyubiquitin protein chain, and induce the target protein to be recognized and degraded by the proteasome. PROTACs are composed of protein targeting ligands, linkers and E3 ubiquitin ligase ligands, which can induce the E3 ligase to approach the target protein to form a target protein-PROTAC-E3 ligase ternary complex. Then the target proteins are degraded by the proteasome. 6, 7
VHL Ligands
Deshaies Raymond's team proposed the concept of PROTACs and reported the first PROTAC based on the Skp1-Cullin-F protein complex polypeptide ligand ( I kappa B alpha phosphopeptide) in 2001. 8 Due to the limited cell permeability and the complex synthesis methods of polypeptide analogues, non-peptide E3 ligase ligands are developed. In the early 2000s, the central hydroxyproline residue in the ALAPYIP recognition sequence of hypoxia-inducible factor-1 (HIF-1α) was comfirmed through the hydrogen bond between HIF-1α and the VHL protein. In 2004, Schneekloth and Crews et al. used the ALAPYIP recognition sequence as the first VHL recruiter for cell-permeable PROTACs to degrade the FK506-binding protein (FKBP12). Then, VHL ligands were further optimized to obtain their small molecule analogs. This lays a strong foundation for the widespread application of PROTACs technology that interferes with proteins at the post-translational level. And it will provide an opportunity to create chemical knockdowns to study protein function without genetic modification. 9
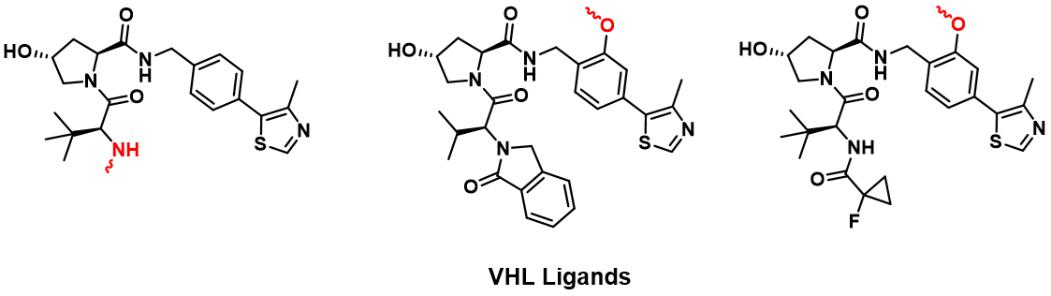
FIG 2: Reported VHL Ligands
CRBN Ligands
During recent decade, other E3 ligase ligands have been discovered, such as Thalidomide. It is a drug originally developed in the 1950s to treat morning sickness in pregnant women. And it was found to have a serious birth defect called seal limb, which is characterized by short limbs. Thalidomide and other immunomodulatory drugs (such as lenalidomide, pomalidomide, etc.) can induce the combination of substrate proteins and CRBN to form a complex, which is mediated by Cullin-RING E3 ubiquitin ligase 4 (CRL4).Then the proteins will be degraded through the ubiquitin-proteasome pathway. Therefore, thalidomide was used for the design and synthesis of PROTACs. 10 In 2019, ARV-110 and ARV-471,which were designed and synthesized based on CRBN ligands, became the first PROTACs to enter clinical trials. In 2020, Arvinas announced interim results from a Phase 1/2 clinical trial of ARV-110 in patients with metastatic castration-resistant prostate cancer (mCRPC). The clinical pharmacological efficacy of PROTACs has been verified, setting a new milestone for the development of PROTACs. 11-13
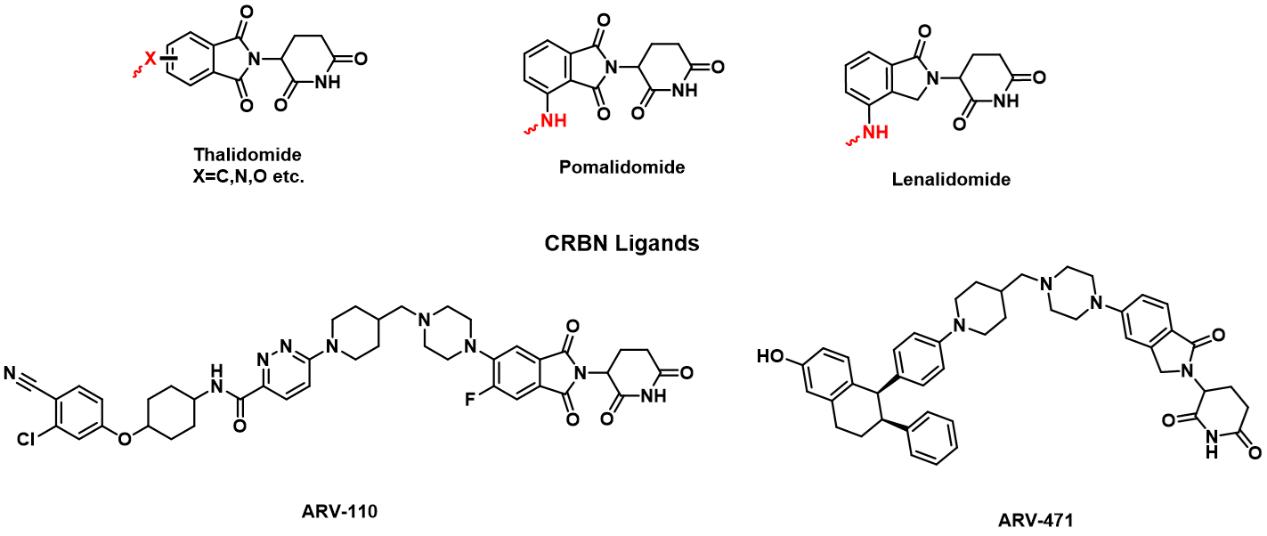
FIG 3: Reported CRBN Ligands and PROTACs in clinical trials.
Other Ligands
Other E3 ligase ligands have been disclosed in recent years, and have been successfully used in the design and synthesis of PROTACs molecules: including mouse double microbody 2 protein (MDM2), apoptosis inhibitory protein (cIAP), RING finger protein 4/114 (RNF4/114), DCAF 11/15/16 (DDB1- and CUL4-associated factor 11/15/16), FEN1B and other E3 ligase ligands. 14, 15
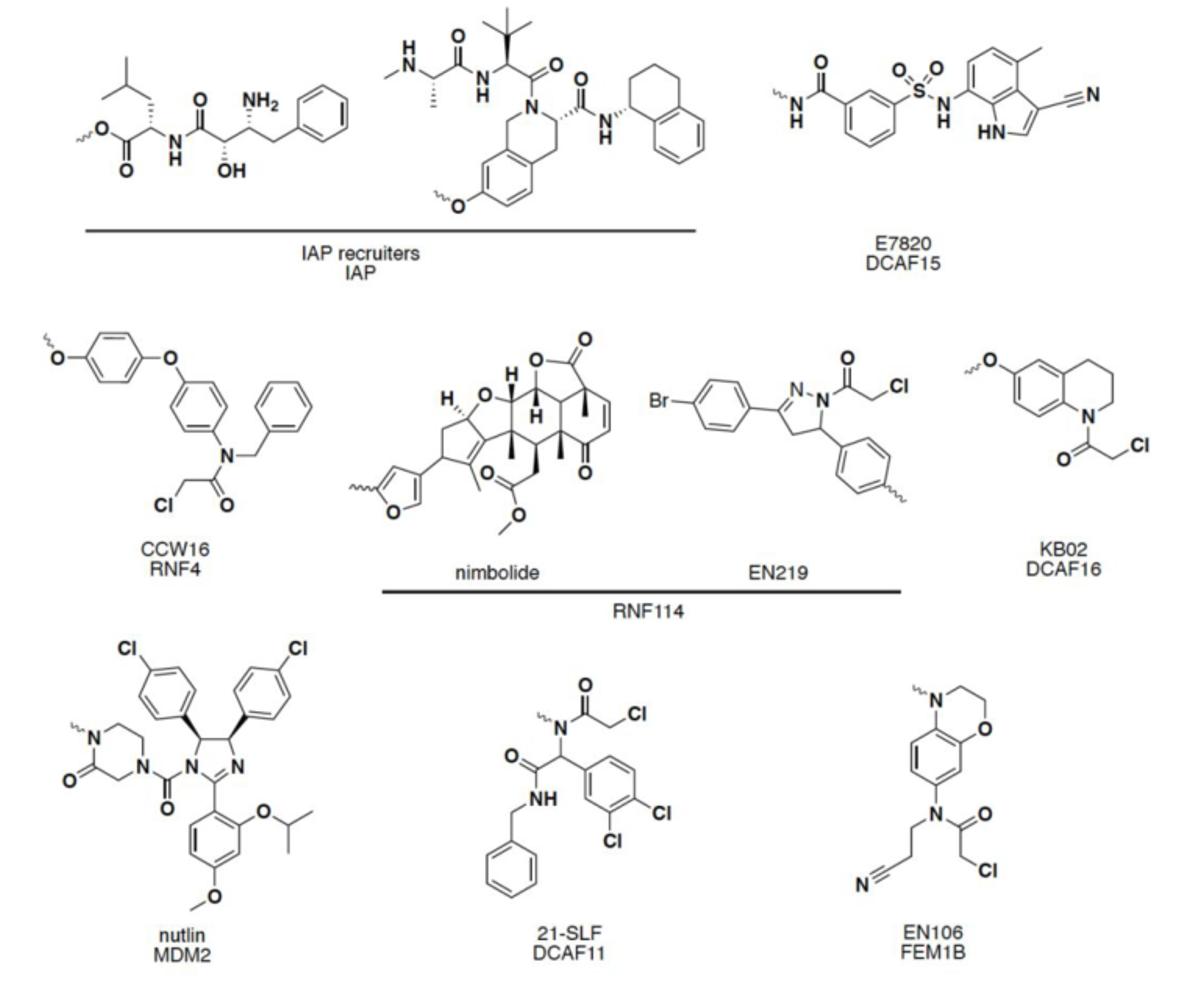
FIG 4: Other ligands
Current E3 ligase recruiters may not be sufficient to degrade any target protein of interest selectively. 16 And compared with more than 600 E3 ligases, only a few E3 ligases have corresponding ligands. The discovery and application of new E3 ligase ligands in PROTACs technology can not only expand the scope of degradable target proteins, but also provide new chemical methods for the treatment of corresponding clinical diseases. 17
References
[1]Ding, Y.; Fei, Y.; Lu, B., Emerging New Concepts of Degrader Technologies. Trends in Pharmacological Sciences 2020, 41, 464-474.
[2]Paiva, S.-L.; Crews, C. M., Targeted protein degradation: elements of PROTAC design. Current Opinion in Chemical Biology 2019, 50, 111-119.
[3]Belcher, B. P.; Ward, C. C.; Nomura, D. K., Ligandability of E3 Ligases for Targeted Protein Degradation Applications. Biochemistry 2021.
[4]Ishida, T.; Ciulli, A., E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS DISCOVERY: Advancing the Science of Drug Discovery 2020, 26, 484-502.
[5]Hanzl, A.; Winter, G. E., Targeted protein degradation: current and future challenges. Current Opinion in Chemical Biology 2020, 56, 35-41.
[6]Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y., PROTACs: great opportunities for academia and industry. Signal Transduction and Targeted Therapy 2019, 4, 64.
[7]Li, X.; Song, Y., Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. Journal of Hematology & Oncology 2020, 13, 50.
[8]Sakamoto Kathleen, M.; Kim Kyung, B.; Kumagai, A.; Mercurio, F.; Crews Craig, M.; Deshaies Raymond, J., Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences 2001, 98, 8554-8559.
[9]Nieto-Jiménez, C.; Morafraile, E. C.; Alonso-Moreno, C.; Ocaña, A., Clinical considerations for the design of PROTACs in cancer. Molecular Cancer 2022, 21, 67.
[10]Kazantsev, A.; Krasavin, M., Ligands for cereblon: 2017–2021 patent overview. Expert Opinion on Therapeutic Patents 2022, 32, 171-190.
[11]Liu, J.; Ma, J.; Liu, Y.; Xia, J.; Li, Y.; Wang, Z. P.; Wei, W., PROTACs: A novel strategy for cancer therapy. Seminars in Cancer Biology 2020, 67, 171-179.
[12]Chamberlain, P. P.; Hamann, L. G., Development of targeted protein degradation therapeutics. Nature Chemical Biology 2019, 15, 937-944.
[13]Wang, C.; Zhang, Y.; Wu, Y.; Xing, D., Developments of CRBN-based PROTACs as potential therapeutic agents. European Journal of Medicinal Chemistry 2021, 225, 113749.
[14]Hughes, G. R.; Dudey, A. P.; Hemmings, A. M.; Chantry, A., Frontiers in PROTACs. Drug Discovery Today 2021, 26, 2377-2383.
[15]Bricelj, A.; Steinebach, C.; Kuchta, R.; Gütschow, M.; Sosič, I., E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points. 2021, 9.
[16]Donovan, K. A.; Ferguson, F. M.; Bushman, J. W.; Eleuteri, N. A.; Bhunia, D.; Ryu, S.; Tan, L.; Shi, K.; Yue, H.; Liu, X.; Dobrovolsky, D.; Jiang, B.; Wang, J.; Hao, M.; You, I.; Teng, M.; Liang, Y.; Hatcher, J.; Li, Z.; Manz, T. D.; Groendyke, B.; Hu, W.; Nam, Y.; Sengupta, S.; Cho, H.; Shin, I.; Agius, M. P.; Ghobrial, I. M.; Ma, M. W.; Che, J.; Buhrlage, S. J.; Sim, T.; Gray, N. S.; Fischer, E. S., Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell 2020, 183, 1714-1731.e10.
[17]Scheepstra, M.; Hekking, K. F. W.; van Hijfte, L.; Folmer, R. H. A., Bivalent Ligands for Protein Degradation in Drug Discovery. Computational and structural biotechnology journal 2019, 17, 160-176.